My Research Projects
These are 3-minute elevator pitches of the research projects that I completed more than 80% of the ground work. For a complete list of publications, here is my google scholar page.
Semi-automated marker gating of Multiplexed Immunofluorescent Images (mIF)
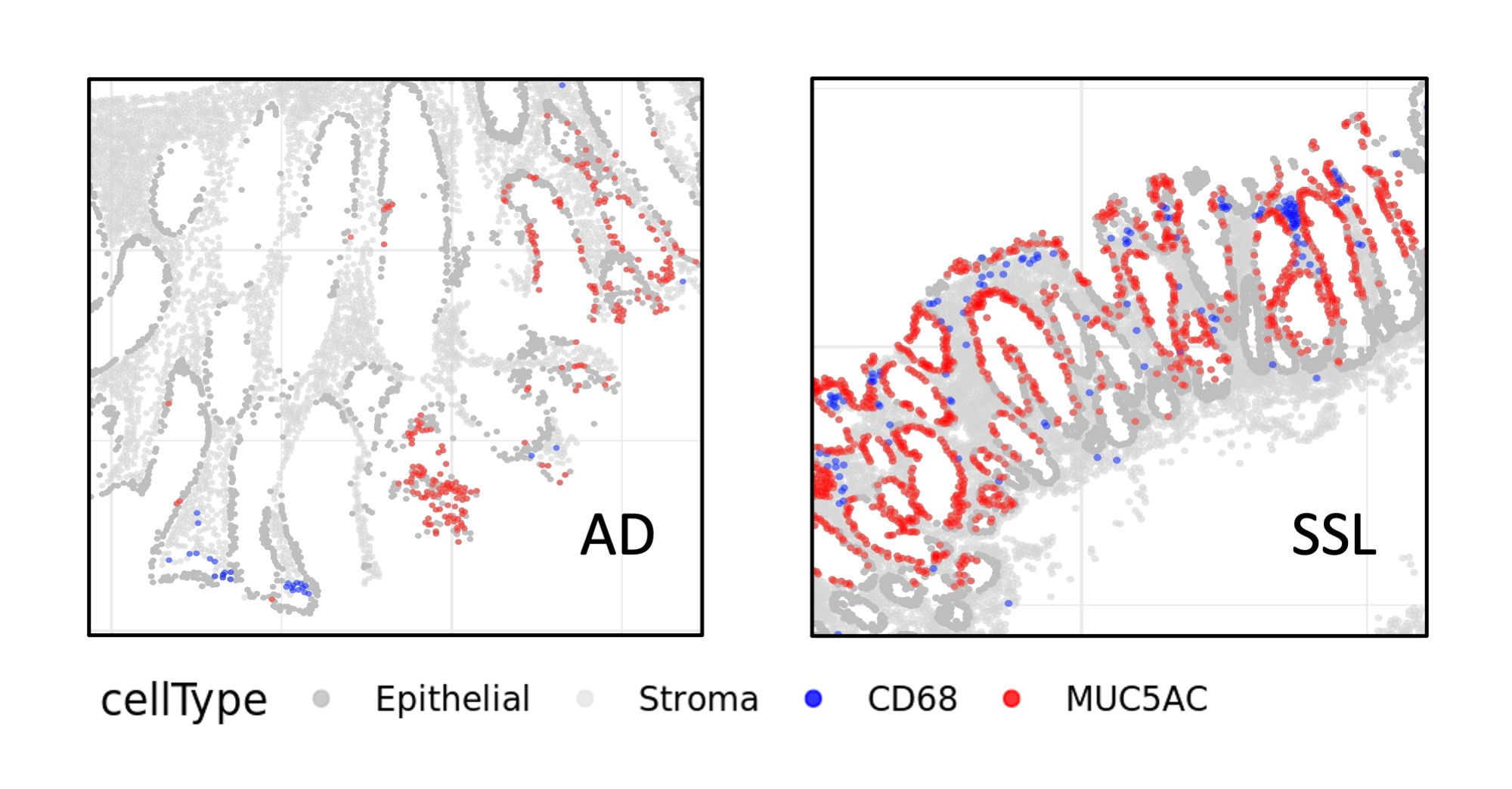
Multiplexed Immunofluorescent (mIF) images is a new and special type of image modality. You can think of mIF as a stack of images, where each images captures different types of marker expression (proteins) of a same tissue. mIF can capture more than 60 different channels and this provides great insight in cell interactions and hereby cancer development pathways. If you happen to read recent ADC (Antibody-drug Conjugate) clinical trial papers, you might find some of them being used as qualitative description of how badly the tissue looks (or how good they are after the treatment).
As a new type of technology, it is still pretty expensive to produce them; the more marker channel with one tissue, the more expensive it is. Therefore, to fully exploit their scientific and monetary values, there has been plenty of studies exploiting the quantitative side of mIF and ended up with great findings.
Of course, with every new data modality there comes “break-in” phases. One of the challenges with processing mIF is marker gating. That is, to identify on each image which marker is expressed for each cell. This could be as easy as a supervised ML classification problem, however with the small sample size and big between sample variances, the result is not often quite ideal.
 Here we proposed using Gamma distribution to build an unsupervised model. We implemented a few changes so that this statistical model is feasible for image data. We implemented the algorithm into an R pacakge (GammaGateR), and we added diagnostic figures (some are interactive, thanks to plotly!) to help quality control the output. You can check out the diagnostic plot in the full vignette.
Here we proposed using Gamma distribution to build an unsupervised model. We implemented a few changes so that this statistical model is feasible for image data. We implemented the algorithm into an R pacakge (GammaGateR), and we added diagnostic figures (some are interactive, thanks to plotly!) to help quality control the output. You can check out the diagnostic plot in the full vignette.
We published a paper in Bioinformatics describing the R package we developed for marker gating in mIF. This paper is featured in NCI.
Public health informatics
This is what I worked on in my masters, and I kept working on similar projects in my spare time now as well: using large language model to uncover information from social media data. I learned this public health research paradigm from excellent Dr. Yulin Hswen. Whenever a major public health event with time sensitivity happens, such as natural disaster, social media is a great resource for learning public opinions on potential damages, and what people want to address the most.
You might recall from your personal experience with Twitter (now X): when you go to the search bar and click on a hashtag, you might be able see several thousands of Tweets. It means that it’s almost impossible to manually process and summarize all of them timely. The great thing is, you can batch process them with topic models such as LDA and BERTopic, which return “topics” of the text inputs. You can also use VADER which is a word-to-emotion dictionary to score the texts regarding their positiveness.
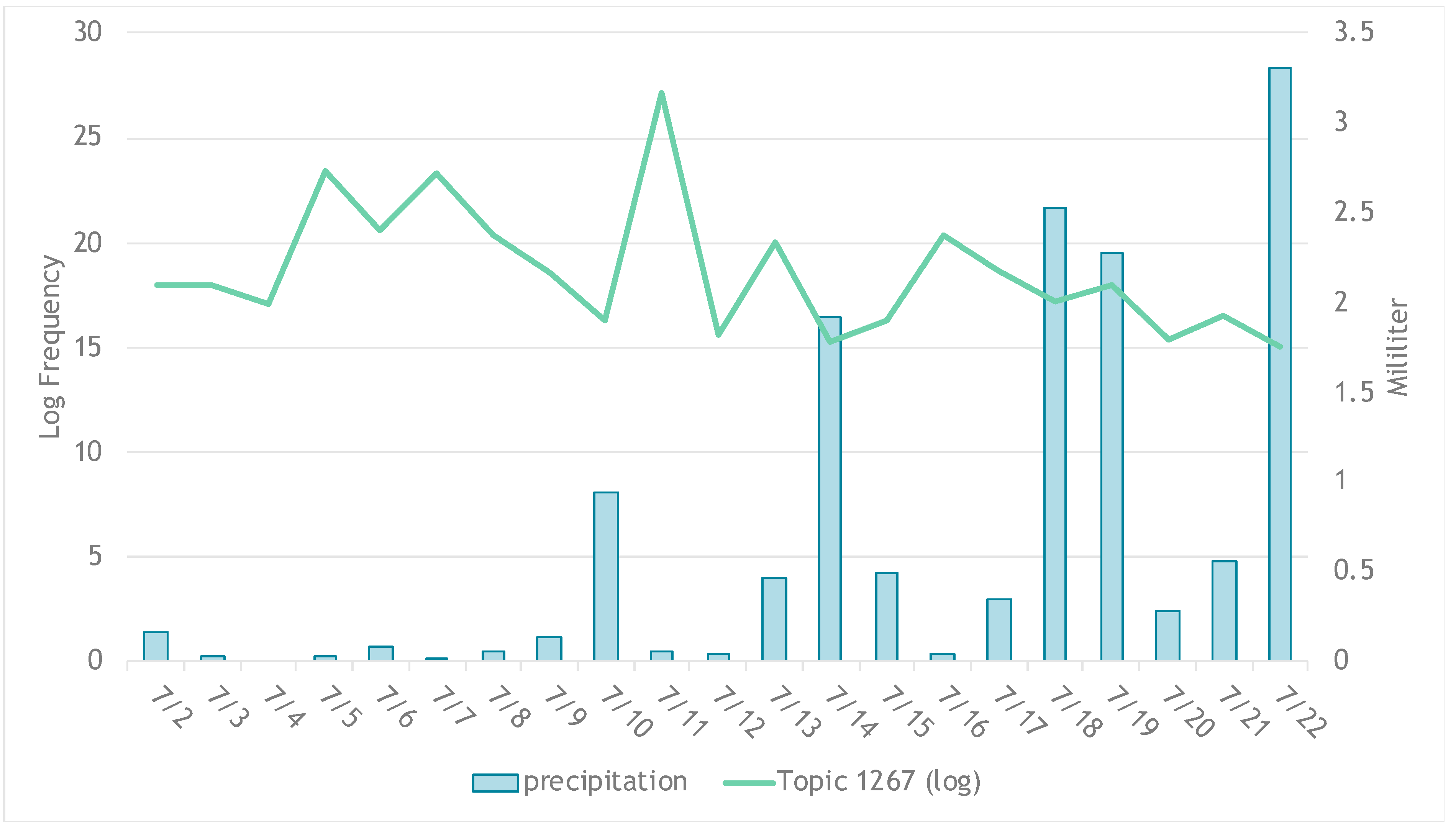 Here comes the scientific question: how do you tell the emotion of each topic, and how much are each topic being discussed? In this paper, I made a little tweak on what we have in the toolbox and quantified similarity of each tweet to find out about tweet-topic assignment and topic discussion frequency. We also observed correlation between rainfall and tweet frequency (for a drought event).
Here comes the scientific question: how do you tell the emotion of each topic, and how much are each topic being discussed? In this paper, I made a little tweak on what we have in the toolbox and quantified similarity of each tweet to find out about tweet-topic assignment and topic discussion frequency. We also observed correlation between rainfall and tweet frequency (for a drought event).
This paper is published in Int. J. Environ. Res. Public Health. I also helped acquiring and processing similar data on a dementia caregiver paper with Dr.Hswen, published in Artificial Intelligence in Health.
Misc
My orals exam documentation is a slightly outdated description of what I aspire to work towards in my current program.